







|
|
|
cDNA Microarray Basics
cDNA microarray technology is considered one of the most important and
powerful tools used to extract and interpret genomic information. The cDNA
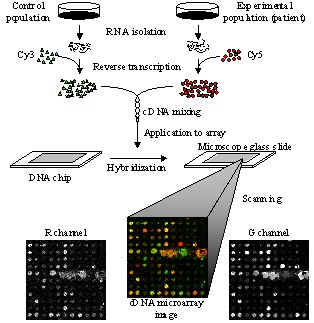
microarray experiment requires to isolate Ribonucleic Acid (RNA) from both
control (known) and experimental (patient) samples. The reverse
transcription process is used to convert the extracted RNAs into cDNAs,
which are further labeled with fluorescent probes, usually Cy3 for the
control and Cy5 for the experimental channel. After subsequent hybridization
and washing procedures, cDNA microarrays are scanned at the ~540 nm (green)
for the control and ~630 nm (red) for the experimental channel respectively.
The scanning procedure produces two 16-bit monochromatic images, which are
further registered into a two-channel, Red-Green image. Analysis of cDNA
microarray data helps in monitoring the expression levels of thousands of
genes simultaneously and provides information relevant to cell activity.
Therefore, cDNA microarrays have found applications in toxicological
research, gene and drug discovery, and disease diagnosis (e.g., cancer,
diabetes, and genetic diseases). |
|
|
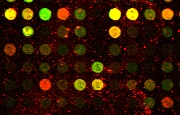 |
The spots of cDNA image constitute
the foreground information which is essential for microarray image
analysis and gene expression tasks. Red spots determine the presence
of RNA from the experimental population of cells, green spots
indicate the presence of RNA from the control population of cells,
and yellow spots determine that RNAs originate from both
experimental and control populations. Arrays of cDNA spots, usually up
to 80 000 probes per 2x4 cm^2 area, are commonly referred to as
microarrays. The vast amount of data and calculations needed to
obtain the relative expression levels of the genes from the
fluorescence intensity at each spot necessitates the development of
automated data processing solutions. |
|
References: |
|
 | R. Lukac and K.N. Plataniotis, "cDNA Microarray
Image Segmentation Using Root Signals," International
Journal of Imaging Systems and Technology, vol. 16, no. 2,
pp. 51-64, April 2006. |
 | R. Lukac, K.N. Plataniotis, B. Smolka, and A.N.
Venetsanopoulos, "cDNA Microarray Image Processing Using Fuzzy
Vector Filtering Framework," Fuzzy Sets and Systems,
Special Issue on Fuzzy Sets and Systems in Bioinformatics,
vol. 152, no. 1, pp.17-35, May 2005. |
 | R. Lukac, B. Smolka, K. Martin, K.N.
Plataniotis, and A.N. Venetsanopoulos, "Vector Filtering for Color
Imaging," IEEE Signal Processing Magazine, Special
Issue on Color Image Processing, vol. 22, no. 1, pp. 74-86, January 2005. |
 | R. Lukac, K.N. Plataniotis, B. Smolka, and A.N.
Venetsanopoulos, "A Multichannel Order-Statistic Technique for
cDNA Microarray Image Processing," IEEE Transactions on NanoBioscience,
vol. 3, no. 4, pp. 272-285, December 2004. |
|
|
|
